Algorithms in PyFibers¶
This page details the logic behind relevant algorithms in PyFibers, including the threshold search and simulation routines. The simulation routines apply the stimulus to the model fiber and track the membrane potential over time. The threshold search determines the minimum stimulus amplitude required to evoke an action potential or block conduction in a model nerve fiber. More details on the ethos behind these algorithms can be found in the PyFibers paper [Marshall et al., 2025].
Threshold Search¶
PyFibers provides find_threshold() to determine the minimum (or “threshold”) stimulus amplitude that yields a specified outcome:
Activation threshold: The lowest amplitude that generates an action potential at a specified node.
Block threshold: The lowest amplitude that prevents propagation of action potentials (conduction block).
Under the hood, find_threshold() executes a bisection search; the method repeatedly calls run_sim() with different stimulus amplitudes until the threshold is bracketed to within a user‐defined tolerance.
Operation of find_threshold()¶
When you call:
amp, ap_info = stimulation.find_threshold(
fiber=my_fiber,
condition="activation", # or "block"
stimamp_top=ub, # upper bound, initial suprathreshold guess
stimamp_bottom=lb, # lower bound, initial subthreshold guess
...,
)
the following steps occur:
Bounds Search
Run simulations at the initial guesses for the upper and lower bounds.
If the upper bound is suprathreshold and the lower bound is subthreshold, proceed to bisection search.
Otherwise, while both bounds are subthreshold or both are suprathreshold, expand the bounds in the appropriate direction.
Repeat until the bounds straddle the threshold, or until the user‐defined maximum number of iterations is reached (default )
Bisection Search
Let
mid = (lb + ub) / 2.Run a simulation at amplitude
mid(by callingrun_sim(mid, fiber)).Determine if it is subthreshold or suprathreshold:
For activation: Suprathreshold if ≥1 action potential is detected (by default, users can change the number of APs required, see
find_threshold()).For block: Suprathreshold if conduction is blocked (i.e., the test action potentials fail to propagate to a distal node).
Update bounds:
If subthreshold → set
lb = mid.If suprathreshold → set
ub = mid.
Repeat until
ub/lbis less than the tolerance (default 1% of amplitude).
Return
The threshold is reported as the upper bound (
ub) once(ub/lb)is small enough to meet the tolerance criteria.
See the figure below for examples of threshold searches with both bounds subthreshold, both bounds suprathreshold, and one where the top bound is suprathreshold and the bottom bound is subthreshold. Below that, a flowchart shows the mechanics of the threshold search algorithm.
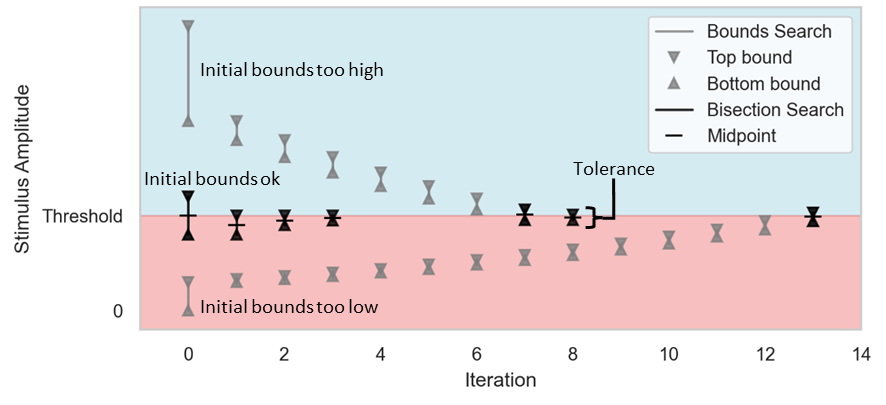
Example threshold searches superimposed on the same axes—one with both initial bounds too low, one with both too high, and one straddling the threshold. For the case with the initial bounds both too low or too high, a bounds search (gray) is first conducted to identify bounds that straddle the threshold, and then the bisection search (black) is initiated. In the case where the initial bounds straddle the threshold, a bisection search begins immediately. The bisection search continues until the difference between the upper and lower bounds is less than the search tolerance, and the threshold is then taken as the upper bound amplitude.¶
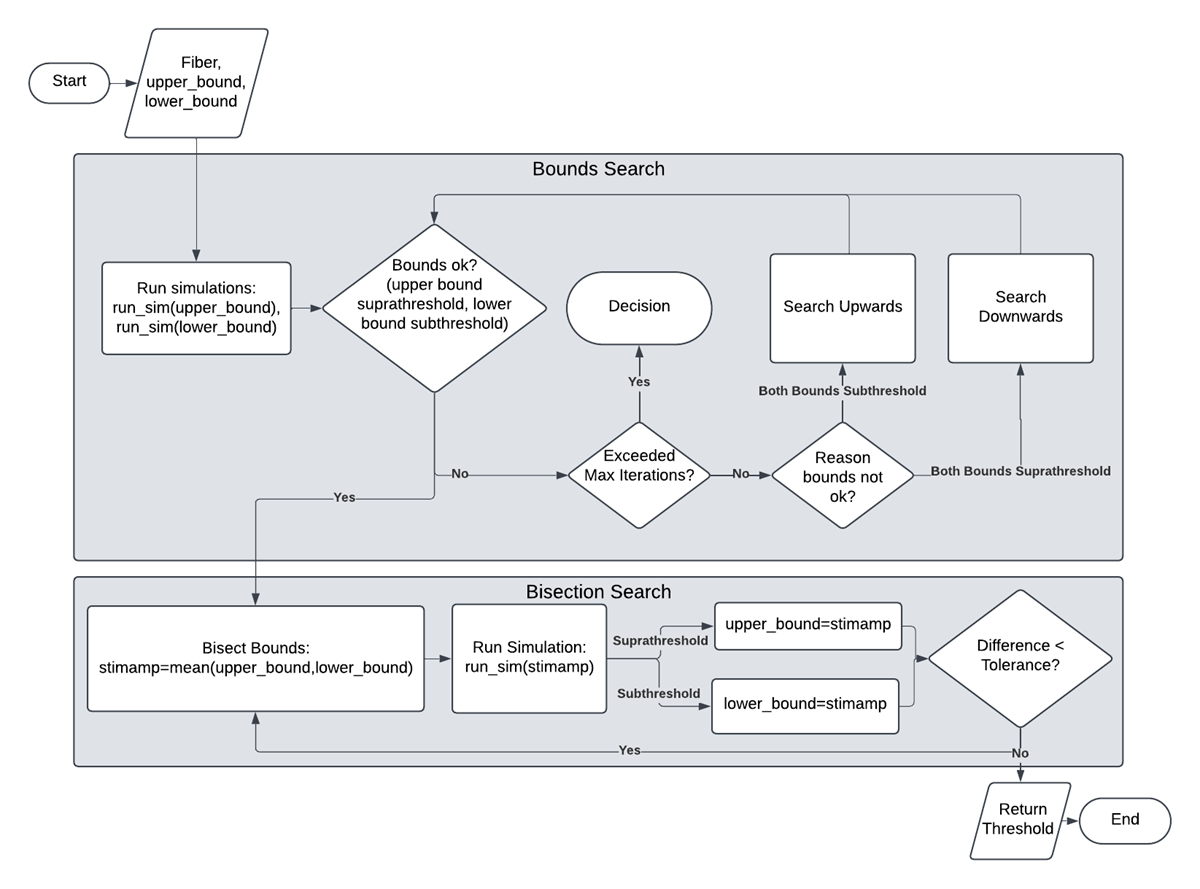
Flowchart of the PyFibers algorithm to identify activation or block threshold. For simplification, several validation checks and details are omitted or simplified. Initial upper and lower bound amplitudes are provided. If the bounds are too low (both subthreshold), an upwards bounds search commences, and if the bounds are too high (both suprathreshold), a downwards bounds search commences. Once the bounds are established (lower bound subthreshold, upper bound suprathreshold), a bisection search executes until the user‐defined exit criterion is reached.¶
Caveats for Block Threshold Searches¶
Warning
While activation threshold is straightforward—did we see an AP?—block threshold can be trickier.
Intrinsic Activity – You must evoke one or more action potentials so that you can test whether conduction is blocked. – PyFibers provides
add_intrinsic_activity()to inject a small depolarizing current at one end, generating ongoing spikes.Re‑excitation at High Amplitude – Kilohertz frequency stimulation can produce re-excitation at high amplitudes, potentially causing a stimulus above the block threshold to appear subthreshold. – In the current implementation, it is up to the user to pick a meaningful upper bound that does not push the fiber into re‑excitation. Future versions may include a more sophisticated block detection algorithm that detects re-excitation, and/or determines the re-excitation threshold in addition to the block threshold.
Onset Response – High‑frequency signals can evoke short‑latency spikes at onset. PyFibers requires a certain delay (
block_delayargument tofind_threshold()) before checking for block.
1.5 Changes That Reduce Threshold Search Runtime¶
Our threshold search was adapted from the algorithm provided in ASCENT [Musselman et al., 2021]. We made several modifications to speed up the search process:
Adaptive Bounds Adjustment – If both initial bounds are subthreshold, the upper bound is raised, but the bottom bound also shifts to keep them straddled more quickly. The reverse applies if both bounds are suprathreshold.
Early Termination for Activation – If activation is the condition, the simulation stops as soon as an action potential is detected in the “detection node.” This can dramatically shorten simulation time for suprathreshold attempts.
Partial Simulation Reuse (Optional) – Once the earliest time point of AP detection is known in an iteration, subsequent runs can skip simulation steps beyond that point plus a user-defined buffer.
These measures collectively can reduce the total simulation time in typical threshold search tasks by >50%, as reported in our manuscript.
run_sim() : Executing Simulations¶
Overview¶
The run_sim() method is the core time-stepped simulation routine in each PyFibers simulation class. It applies the specified stimulation to the model fiber at each time step and uses NEURON’s solver to update the membrane potential and gating variables. At the end, run_sim() returns:
The number of action potentials recorded at a designated node (by default, the one closest to 90% fiber length).
The time of the last action potential crossing (if any).
When find_threshold() is called, it repeatedly calls:
run_sim(mid_amplitude, fiber)
Therefore, custom run_sim() implementations should follow these constraints. For more information, see building custom simulations.
ScaledStim.run_sim()¶
ScaledStim is designed for extracellular stimulation. It expects that the fiber has an array of extracellular potentials (e.g., from a point source), each scaled by the provided stimulus amplitude and waveform(s) at each time step. The process is detailed below, and summarized in the diagram below.
For each source, given the spatial distribution of extracellular potentials at the center of each fiber section:
and the waveform at each time point:
the dot product gives the unscaled extracellular potential at each fiber section and each time point:
which can then be scaled by the desired stimulation amplitude (a):
Under the principle of linearity, the extracellular potentials from multiple sources ((m)) are summed:
The final matrix of potentials is applied to each section at each time step.
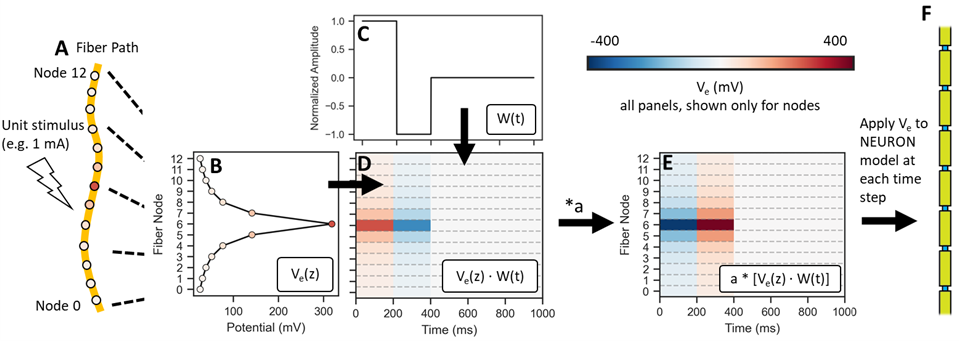
Process of calculating the spatiotemporal profile of extracellular potentials applied to the model fiber by run_sim(), which incorporates the spatial distribution of potentials in response to 1 mA stimulation (\(V_e(z)\)), the unit waveform (\(W(t)\)), and the stimulation amplitude (\(a\)). If potentials from multiple sources and the corresponding waveforms are provided, steps A–D are performed for each combination of source/waveform/stimulation amplitude, and the results are summed across sources before proceeding.¶
IntraStim.run_sim()¶
IntraStim is a simpler class for intracellular stimulation. It injects current directly into a chosen fiber section. Key points:
The user specifies:
Pulse parameters: width, frequency, duration.
Location along the fiber: e.g., the middle node or the first node.
IntraStim.run_sim(amplitude, fiber)sets up and applies a square current pulse inside the specified section. The amplitude is scaled by the given stimulation amplitude.For multi‑pulse waveforms,
IntraStimrepeats square pulses at intervals (pulse repetition frequency) untiltstop.