Custom Fiber Models¶
Creating a new fiber model involves defining a subclass of Fiber and implementing methods that describe the specific mechanisms and ultrastructure of the fiber. This section provides a step-by-step guide on how to create a new fiber model.
Note
There are two main ways to make your custom fiber model available in PyFibers:
Runtime Registration (Recommended for development/testing): Use the
register_custom_fiber()function to register your fiber model at runtime. This allows you to usebuild_fiberwith your custom model without modifying the main package.Plugin System (Recommended for distribution): Create a separate package and make it discoverable as a plugin. This is the best approach for sharing your fiber model with others.
For most users, we recommend starting with Runtime Registration for development and testing, then moving to the Plugin System for distribution.
The implementation of a custom fiber model requires custom .MOD file(s) that describe the membrane mechanisms (e.g., ion channels) of your model, as well as a Python class that defines the fiber model. Note that models can either be homogeneous (each section is identical, e.g., Hodgkin-Huxley) or heterogeneous (sections can vary in repeating patterns, e.g., MRG). We will start with a homogeneous fiber model, and then describe how to extend the methods to a heterogeneous model.
Steps to Create a Fiber Model Subclass¶
Let’s walk through creating a custom fiber model step by step, using a Hodgkin-Huxley (homogeneous) fiber as an example:
Step 1: Inherit from Fiber¶
Your new class should inherit from Fiber:
import neuron
from neuron import h
from pyfibers import Fiber, FiberModel, build_fiber, register_custom_fiber
h.load_file("stdrun.hoc")
class HHFiber(Fiber):
"""Hodgkin-Huxley fiber model."""
Step 2: Specify the Submodels¶
Define the submodels attribute as a list of the submodels that your fiber model uses. Often, this list will contain only a single item. Each item in the list should be capitalized and contain only letters and underscores; these strings are used to define the name of each fiber model as accessed from the FiberModel enum (for example, one would access the fiber model by calling FiberModel.MY_HOMOGENEOUS_FIBER_MODEL). If your class has multiple subtypes (such as in {py:class}~pyfibers.models.mrg.MRGFiber), you can define multiple submodels. When you create an instance of your fiber model, it will gain the enum as the self.fiber_model attribute. Check the name by accessing self.fiber_model.name.
class HHFiber(Fiber):
"""Hodgkin-Huxley fiber model."""
submodels = ["HH"] # This will be available as FiberModel.HH
Step 3: Initialize the Subclass¶
Define the __init__ method, call the superclass initializer, and set any model-specific parameters. At minimum, set self.v_rest (resting membrane potential) and self.myelinated (whether the fiber is myelinated). It is also recommended to specify gating variables if you want to be able to record these values during simulations (these are specified in the .mod files describing the node mechanisms).
For homogeneous fiber models, set the delta_z parameter, which is the distance from the center of one node to the next node. This can be passed as an argument to __init__.
def __init__(self, diameter: float, delta_z: float = 8.333, **kwargs):
"""Initialize HHFiber class."""
super().__init__(diameter=diameter, **kwargs)
self.gating_variables = {
"h": "h_hh",
"m": "m_hh",
"n": "n_hh",
}
self.myelinated = False
self.delta_z = delta_z
self.v_rest = -65 # mV
For heterogeneous fiber models (e.g., myelinated fibers with multiple section types), delta_z must be calculated by you (the model implementer) in your __init__ method based on the fiber diameter. It should not be accepted as an argument from users. You must:
Check that
delta_zis not inkwargsand raise an error if it isCalculate and set
self.delta_zin your__init__method based on the diameter (or from model-specific parameters)
def __init__(self, diameter: float, **kwargs):
"""Initialize heterogeneous fiber class."""
if "delta_z" in kwargs:
raise ValueError("Cannot specify delta_z for this fiber model")
super().__init__(diameter=diameter, **kwargs)
# You must calculate delta_z based on diameter
self.delta_z = self.diameter * 100 # Example: simple calculation
Step 4: Define the Node Creation Method(s)¶
Implement method(s) that create the specific sections of the fiber model. For a homogeneous fiber model, you will create a single method. For a heterogeneous fiber model, you will create multiple methods. These methods should return a NEURON h.Section object representing the node or section.
Note
To incorporate custom mechanisms into the section method, you should place the .mod files in a directory, compile them using nrnivmodl, and then load them by placing neuron.load_mechanisms(dir) at the top of your python file, where “dir” is the directory containing your compile mechanisms.
def create_hh(self, ind: int, node_type: str):
"""Create a Hodgkin-Huxley node."""
node = h.Section(name=f"{node_type} node {ind}") # create section
node.L = self.delta_z # length of node
node.diam = self.diameter # diameter of fiber
node.nseg = 1 # one segment
node.insert("extracellular") # extracellular NEURON mechanism
node.xc[0] = 0 # short circuit
node.xg[0] = 1e10 # short circuit
node.v = self.v_rest # rest potential
node.insert("hh") # hodgkin-huxley mechanism built into NEURON
node.Ra = 100 # axial resistivity
node.cm = 1 # membrane capacitance
return node
Step 5: Define the generate Method¶
Implement the generate method, which calls the superclass generate() method with a list of functions that create the specific sections of the fiber model. For a homogeneous fiber model, you will pass a single function in the list. For a heterogeneous fiber model, you will pass a list of functions.
def generate(self, **kwargs):
"""Generate the fiber model sections."""
return super().generate([self.create_hh], **kwargs)
Runtime Registration (Recommended for Development)¶
The easiest way to use your custom fiber model is to register it at runtime using the register_custom_fiber() function. This approach allows you to use your custom fiber with the standard build_fiber function and FiberModel enum without modifying the main PyFibers package.
Basic Usage¶
from pyfibers import FiberModel, build_fiber, register_custom_fiber
from my_custom_fiber import MyCustomFiber
# Register your custom fiber model
register_custom_fiber(MyCustomFiber)
# Now you can use it with build_fiber
model = FiberModel.MY_CUSTOM_FIBER # Uses the submodels attribute
fiber = build_fiber(diameter=5.7, fiber_model=model, temperature=37, n_nodes=21)
Complete Example¶
Here’s the complete HHFiber class definition:
"""Example: Creating and using a custom Hodgkin-Huxley fiber model."""
import neuron
from neuron import h
from pyfibers import Fiber, FiberModel, build_fiber, register_custom_fiber
h.load_file("stdrun.hoc")
class HHFiber(Fiber):
"""Hodgkin-Huxley fiber model."""
submodels = ["HH"] # This will be available as FiberModel.HH
def __init__(self, diameter: float, delta_z: float = 8.333, **kwargs):
"""Initialize HHFiber class."""
super().__init__(diameter=diameter, **kwargs)
self.gating_variables = {
"h": "h_hh",
"m": "m_hh",
"n": "n_hh",
}
self.myelinated = False
self.delta_z = delta_z
self.v_rest = -65 # mV
def generate(self, **kwargs):
"""Generate the fiber model sections."""
return super().generate([self.create_hh], **kwargs)
def create_hh(self, ind: int, node_type: str):
"""Create a Hodgkin-Huxley node."""
node = h.Section(name=f"{node_type} node {ind}") # create section
node.L = self.delta_z # length of node
node.diam = self.diameter # diameter of fiber
node.nseg = 1 # one segment
node.insert("extracellular") # extracellular NEURON mechanism
node.xc[0] = 0 # short circuit
node.xg[0] = 1e10 # short circuit
node.v = self.v_rest # rest potential
node.insert("hh") # hodgkin-huxley mechanism built into NEURON
node.Ra = 100 # axial resistivity
node.cm = 1 # membrane capacitance
return node
Heterogeneous Fiber Models¶
For heterogeneous fiber models (e.g., myelinated fibers with nodes and myelin sections), the process is similar but you would define multiple creation methods and pass them as a list to the generate method:
def generate(self, **kwargs):
"""Generate the fiber model sections."""
function_list = [
self.create_node,
self.create_myelin,
]
return super().generate(function_list, **kwargs)
Homogeneous vs Heterogeneous Fiber Models¶
The class construction will differ depending on whether you are creating a “homogeneous” fiber model (i.e., all sections of the fiber are identical, typically unmyelinated fibers) or a “heterogeneous” fiber model (i.e., sections of the fiber have different properties, such as nodes and myelin). See the figure below for a visual representation of the construction of homogeneous and heterogeneous fiber models.
Tip
Reference implementations: You can find complete examples of both homogeneous and heterogeneous fiber models in the PyFibers codebase:
Homogeneous models: See
pyfibers.models.sundt,pyfibers.models.schild,pyfibers.models.rattay,pyfibers.models.thio, andpyfibers.models.tigerholmHeterogeneous models: See
pyfibers.models.mrgandpyfibers.models.sweeney
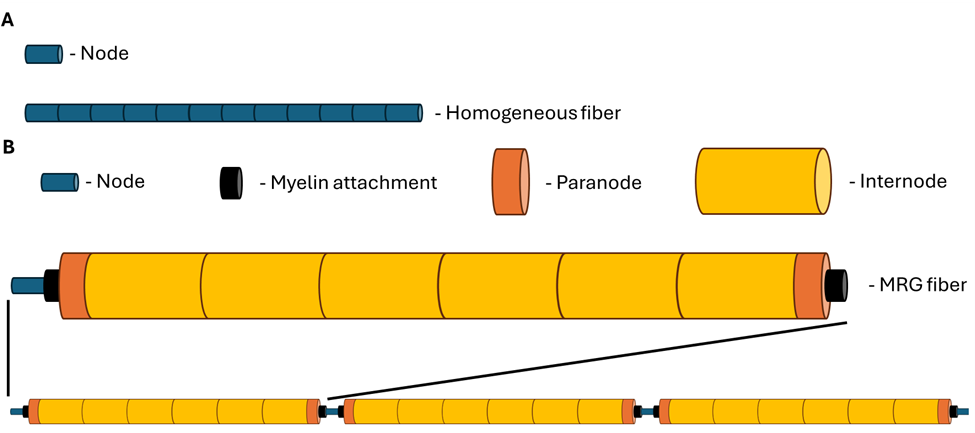
Construction of a model fiber. A) For unmyelinated fibers and myelinated fibers with only one section type (i.e., homogeneous fiber), a fiber model describes a single set of mechanisms and ultrastructure for a node, which is then repeated to the target number of sections. B) For fibers with multiple section types (e.g., MRG as shown), a repeating series of sections with heterogeneous membrane mechanisms and ultrastructure is described from one node until just before the next node. This sequence is repeated to one less than the target number of nodes, and a node is added to the end of the fiber for symmetry. Example shown: MRG model fiber with 4 nodes and 11 sections per node, resulting in a final section count of nsections = (nnodes - 1) * 11 + 1 = (4 - 1) * 11 + 1 = 34. Figure is not to scale.¶
Plugin System (Recommended for Distribution)¶
For sharing your fiber model with others or creating a permanent, distributable package, the plugin system is the best approach. Plugins are automatically discovered when PyFibers is imported and provide a clean way to extend the framework.
Creating a Plugin Package¶
Other research groups may wish to create their own fiber models in the PyFibers environment and publish them as a separate public repository. Such fiber models can be made discoverable as plugins which will become automatically available in PyFibers after installation. To make your fiber model discoverable as a plugin, follow these steps:
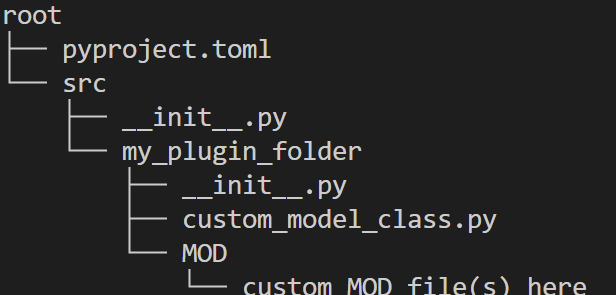
Create a Python package for your fiber model: Create a Python package for your fiber model and include the necessary files and classes. We recommend following Python’s packaging tutorial. An example directory structure for a fiber model package is shown below:
Specify the Entry Point for Your Fiber Model: The entry point tells
PyFibersthat your fiber model should be imported and where to find the class that defines it. Entry points can be specified in multiple ways; here we provide an example using pyproject.toml:
[project.entry-points."pyfibers.fiber_plugins"]
my_fiber_model = "my_plugin_folder.custom_model_class:MyFiberModelClass"
Install your package and verify that it works with
PyFibers. Upon installation, your fiber model should be automatically discovered and available for use inPyFibers. The necessary MOD files should be compiled automatically whenPyFibersis imported.Publish Your Fiber Model Package: Publish your fiber model package to a repository such as PyPI. Once published, other users can install your fiber model package using pip. After installation, your fiber model will be automatically discovered and available for use in
PyFibers. The name of the fiber model will be given by thesubmodelsattribute of your class (see the instructions for creating fiber models above).