Basic simulation and search for activation threshold¶
This tutorial will use the fiber model created in the fiber creation tutorial to run a basic simulation of fiber stimulation. Then we will demonstrate running a bisection search for the fiber’s activation threshold (i.e., the minimum stimulation amplitude needed to generate an action potential).
Create the fiber¶
As in the fiber creation tutorial, we use build_fiber() to create a 10 µm diameter MRG fiber.
from pyfibers import build_fiber, FiberModel
# create fiber model
n_nodes = 25
fiber = build_fiber(FiberModel.MRG_INTERPOLATION, diameter=10, n_nodes=n_nodes)
import pyfibers
# Enable INFO level logging to see simulation progress
pyfibers.enable_logging()
Simulation setup¶
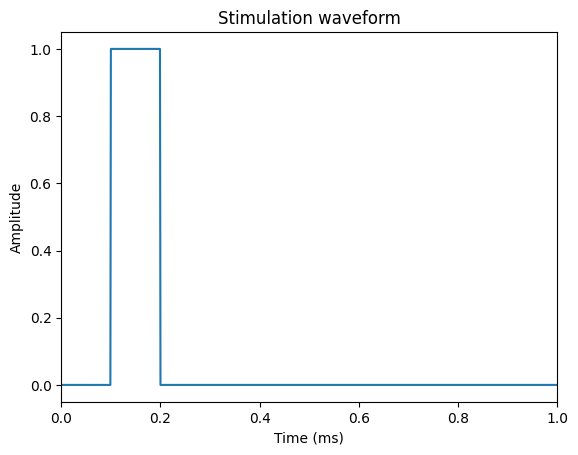
Before we can run a simulation, we need to create a stimulation waveform (i.e., I(t), the time‐course of the extracellular stimulation). We use a biphasic rectangular pulse in this tutorial. We also specify our simulation parameters as constants.
See documentation on stimulation waveforms for more information on creating different waveforms.
from scipy.interpolate import interp1d
time_step = 0.001 # milliseconds
time_stop = 20 # milliseconds
start, on, off = 0, 0.1, 0.2 # milliseconds
waveform = interp1d(
[start, on, off, time_stop], [0, 1, 0, 0], kind="previous"
) # monophasic rectangular pulse
Plot the waveform to see what it looks like.
import matplotlib.pyplot as plt
import numpy as np
time_steps = np.arange(0, time_stop, time_step)
plt.plot(time_steps, waveform(time_steps))
plt.xlim(0, 1)
plt.title('Stimulation waveform')
plt.xlabel('Time (ms)')
plt.ylabel('Amplitude')
plt.show()
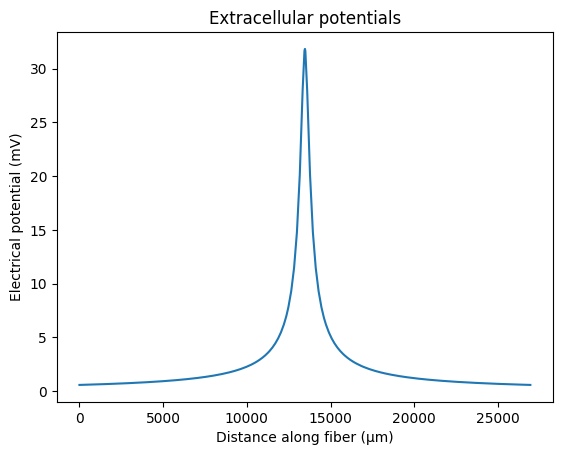
We also need extracellular potentials along the length of the fiber. We will use an extracellular point source for this tutorial, 250 µm from the fiber, positioned over its center. Our fiber has 265 sections, so we need 265 potentials (i.e., one for the middle of each section). Typically, extracellular potentials are generated with a “unit” current source (i.e., 1 mA).
In many cases, users will want to use electrical potentials generated using an outside source (e.g., a finite element model). See our documentation on extracellular potentials for more information.
fiber.potentials = fiber.point_source_potentials(0, 250, fiber.length / 2, 1, 10)
plt.plot(fiber.longitudinal_coordinates, fiber.potentials)
plt.xlabel('Distance along fiber (µm)')
plt.ylabel('Electrical potential (mV)')
plt.title('Extracellular potentials')
plt.show()
Running a simulation¶
To run a simulation, we will use an instance of the ScaledStim class. This class is used to run simulations of a fiber’s response to extracellular stimulation. For more information on parameters, see the ScaledStim Documentation.
from pyfibers import ScaledStim
# Create instance of ScaledStim class
stimulation = ScaledStim(waveform=waveform, dt=time_step, tstop=time_stop)
print(stimulation)
ScaledStim: 20 ms (dt=0.001 ms) (t_init_ss=-200 ms, dt_init_ss=5 ms)
We can use the run_sim() method of the ScaledStim class to run a simulation. By default, this method monitors for action potentials at the distal end of the fiber. It returns the number of detected action potentials and the time of the last action potential. Since we used a unit current source (1 mA), our stimamp (stimulation amplitude) here represents the current of the stimulus (in mA).
stimamp = -1.5 # technically unitless, but scales the unit (1 mA) stimulus to 1.5 mA
ap, time = stimulation.run_sim(stimamp, fiber)
print(f'Number of action potentials detected: {ap}')
print(f'Time of last action potential detection: {time} ms')
INFO:pyfibers.stimulation:Running: -1.5
INFO:pyfibers.stimulation:N aps: 1, time 0.379
Number of action potentials detected: 1.0
Time of last action potential detection: 0.379 ms
Run search for activation threshold¶
If we want to determine the activation threshold, we can use the find_threshold() method.
This method returns the stimulation amplitude at which the fiber begins to activate and the number of generated action potentials. The threshold amplitude is calculated using a bisection search (see the Documentation on Algorithms in PyFibers).
amp, ap = stimulation.find_threshold(fiber)
print(f'Activation threshold: {amp} mA')
INFO:pyfibers.stimulation:Running: -1
INFO:pyfibers.stimulation:N aps: 1, time 0.437
INFO:pyfibers.stimulation:Running: -0.01
INFO:pyfibers.stimulation:N aps: 0, time None
INFO:pyfibers.stimulation:Search bounds: top=-1, bottom=-0.01
INFO:pyfibers.stimulation:Found AP at 0.437 ms, subsequent runs will exit at 5.437 ms. Change 'exit_t_shift' to modify this.
INFO:pyfibers.stimulation:Beginning bisection search
INFO:pyfibers.stimulation:Search bounds: top=-1, bottom=-0.01
INFO:pyfibers.stimulation:Running: -0.505
INFO:pyfibers.stimulation:N aps: 0, time None
INFO:pyfibers.stimulation:Search bounds: top=-1, bottom=-0.505
INFO:pyfibers.stimulation:Running: -0.7525
INFO:pyfibers.stimulation:N aps: 0, time None
INFO:pyfibers.stimulation:Search bounds: top=-1, bottom=-0.7525
INFO:pyfibers.stimulation:Running: -0.87625
INFO:pyfibers.stimulation:N aps: 0, time None
INFO:pyfibers.stimulation:Search bounds: top=-1, bottom=-0.87625
INFO:pyfibers.stimulation:Running: -0.938125
INFO:pyfibers.stimulation:N aps: 0, time None
INFO:pyfibers.stimulation:Search bounds: top=-1, bottom=-0.938125
INFO:pyfibers.stimulation:Running: -0.969062
INFO:pyfibers.stimulation:N aps: 1, time 0.477
INFO:pyfibers.stimulation:Search bounds: top=-0.969062, bottom=-0.938125
INFO:pyfibers.stimulation:Running: -0.953594
INFO:pyfibers.stimulation:N aps: 1, time 0.548
INFO:pyfibers.stimulation:Search bounds: top=-0.953594, bottom=-0.938125
INFO:pyfibers.stimulation:Running: -0.945859
INFO:pyfibers.stimulation:N aps: 0, time None
INFO:pyfibers.stimulation:Search bounds: top=-0.953594, bottom=-0.945859
INFO:pyfibers.stimulation:Running: -0.949727
INFO:pyfibers.stimulation:N aps: 1, time 0.609
INFO:pyfibers.stimulation:Threshold found at stimamp = -0.949727
INFO:pyfibers.stimulation:Validating threshold...
INFO:pyfibers.stimulation:Running: -0.949727
INFO:pyfibers.stimulation:N aps: 1, time 0.609
Activation threshold: -0.9497265625 mA
Intracellular stimulation¶
We can also use intracellular stimulation by using the IntraStim class.
from pyfibers import IntraStim
# reset fiber potentials to zero to eliminate extracellular stimulation
fiber.potentials = np.zeros(len(fiber.coordinates))
# Intracellular stimulation arguments
clamp_kws = {
'delay': 1.0, # milliseconds
'pw': 0.1, # milliseconds, pulse width per pulse
'dur': 10, # milliseconds, total duration of stimulation
'freq': 1000, # pulse repetition frequency in Hz
'amp': 1, # nA, intracellular current amplitude. We'll set to 1 and scale with run_sim
}
loc = 0.5 # location of the intracellular electrode along the fiber
stimulation = IntraStim(
istim_loc=loc, dt=time_step, tstop=time_stop, clamp_kws=clamp_kws
)
# Run simulation with intracellular stimulation
stimamp = 2 # nA, positive since stimulation is intracellular
ap, time = stimulation.run_sim(stimamp, fiber)
print(f'Number of action potentials detected: {ap}')
print(f'Time of last action potential detection: {time} ms')
INFO:pyfibers.stimulation:Running: 2
INFO:pyfibers.stimulation:N aps: 5, time 9.396
Number of action potentials detected: 5.0
Time of last action potential detection: 9.396 ms
As before, we can run a search for activation threshold.
amp, ap = stimulation.find_threshold(fiber, stimamp_top=1, stimamp_bottom=0)
print(f'Activation threshold: {amp} nA')
INFO:pyfibers.stimulation:Running: 1
INFO:pyfibers.stimulation:N aps: 1, time 1.424
INFO:pyfibers.stimulation:Running: 0
INFO:pyfibers.stimulation:N aps: 0, time None
INFO:pyfibers.stimulation:Search bounds: top=1, bottom=0
INFO:pyfibers.stimulation:Found AP at 1.424 ms, subsequent runs will exit at 6.4239999999999995 ms. Change 'exit_t_shift' to modify this.
INFO:pyfibers.stimulation:Beginning bisection search
INFO:pyfibers.stimulation:Search bounds: top=1, bottom=0
INFO:pyfibers.stimulation:Running: 0.5
INFO:pyfibers.stimulation:N aps: 0, time None
INFO:pyfibers.stimulation:Search bounds: top=1, bottom=0.5
INFO:pyfibers.stimulation:Running: 0.75
INFO:pyfibers.stimulation:N aps: 0, time None
INFO:pyfibers.stimulation:Search bounds: top=1, bottom=0.75
INFO:pyfibers.stimulation:Running: 0.875
INFO:pyfibers.stimulation:N aps: 0, time None
INFO:pyfibers.stimulation:Search bounds: top=1, bottom=0.875
INFO:pyfibers.stimulation:Running: 0.9375
INFO:pyfibers.stimulation:N aps: 0, time None
INFO:pyfibers.stimulation:Search bounds: top=1, bottom=0.9375
INFO:pyfibers.stimulation:Running: 0.96875
INFO:pyfibers.stimulation:N aps: 0, time None
INFO:pyfibers.stimulation:Search bounds: top=1, bottom=0.96875
INFO:pyfibers.stimulation:Running: 0.984375
INFO:pyfibers.stimulation:N aps: 1, time 1.485
INFO:pyfibers.stimulation:Search bounds: top=0.984375, bottom=0.96875
INFO:pyfibers.stimulation:Running: 0.976562
INFO:pyfibers.stimulation:N aps: 1, time 1.562
INFO:pyfibers.stimulation:Search bounds: top=0.976562, bottom=0.96875
INFO:pyfibers.stimulation:Running: 0.972656
INFO:pyfibers.stimulation:N aps: 1, time 1.681
INFO:pyfibers.stimulation:Threshold found at stimamp = 0.972656
INFO:pyfibers.stimulation:Validating threshold...
INFO:pyfibers.stimulation:Running: 0.972656
INFO:pyfibers.stimulation:N aps: 3, time 7.472
Activation threshold: 0.97265625 nA
See also the tutorial for analyzing results.